The availability of a mutant line in which a single gene has been disrupted gives biologists a powerful
tool in understanding the function of that gene. Thus, sequence-indexed collections of single
insertions are critical resources for elucidating gene function in organisms with sequenced genomes.
Our approach to creating such a resource in maize entails generating, sequencing, and indexing new
insertions of the transposons Ds and Ac, which tend to transpose to nearby sites.
Under funding from the NSF Plant Genome Program, we have produced a set of around 100 T-DNA launching platforms carrying Ds*
elements marked with GFP. Our system allows simple visual selection of element transposition from
many different regions of the genome and, thus, enables researchers to generate regional gene knock-out
collections. We have recently published a detailed step-by-step protocol for generating such a
collection from any one of our Ds* launching platforms (Li et al. 2013). In addition, using
a combination of next-generation sequencing technology, 3-dimensional pooling, and a software package
specifically developed for this project (Xiong et al., 2013), we are generating a sequence-indexed
collection of 14,000 Ds* insertions scattered around the genome. All insertion site
sequences are searchable in our database and in maize GDB, where they are cross-referenced to stocks
available from the Maize Genetics Stock Center.
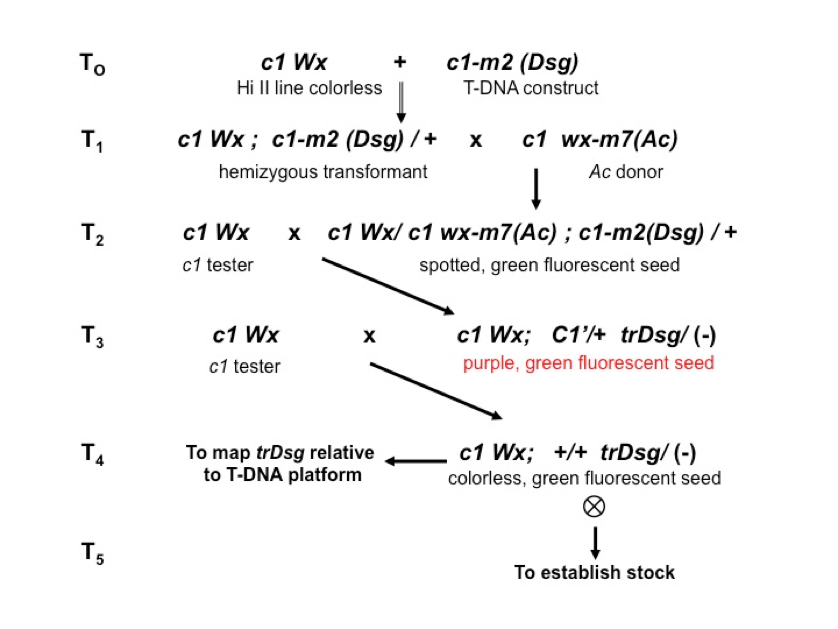
Our T-DNA platforms carry the c1-m(Dsg) engineered reporter allele
(T0) and produce a green fluorescent, Ac-dependent spotted
phenotype (T2). In our system, Ac activity is provided
by the native wx-m7(Ac) allele on chromosome 9
(T1). From any given platform, we select transpostions of
Dsg (T3) as exceptional purple revertants that retain green
fluorescence (C’+Dsg), so the spotted phenotype is lost at the
outset. We cross those selections to a c1 tester to confirm the
concordancy of revertants (i.e., that the selection is heritable) and to
determine the linkage between the transposed Dsg (trDsg) and
the corresponding T-DNA platforms (T3). In the case of unlinked
or loosely linked transpositions, we select from the testcross progeny
colorless (c), green fluorescent (Dsg) kernels (T4) for
sequence-indexing and for seed propagation. If the trDsg is so
closely linked that it produces no or very few colorless, green fluorescent seed
in the testcross progeny, we select purple (C’) green fluorescent
(Dsg) kernels for sequence-indexing and for seed propagation, but these
are rare among materials sent to the Co-op. We deposit all
sequence-indexed selfed progenies (T5) in the Co-op.
What recipients of our materials need to ensure is that they work with green
fluorescent seed, as the Dsg element is segregating in the progeny.
We do not select for or against wx. If the waxy endosperm
phenotype is still segregating, selecting for it should retain Ac, in
case the researcher wants to initiate secondary movements of the mapped
trDsg. Ac can transpose from wx-m7(Ac) leaving
a defective wx allele, but such transpositions are rare. Selecting against
wx should eliminate Ac, unless Ac transposed from
wx during the derivation of the trDsg stock. Presence or
absence of Ac can be definitively ascertained by crossing to the native
c1-m2 allele (available from the Co-op)
